MedRecord#
1. Preface#
Every major library has a central object that constitutes its core. For PyTorch, it is the torch.Tensor, whereas for Numpy, it is the np.array. In our case, MedModels centres around the MedRecord as its foundational structure.
MedModels delivers advanced data analytics methods out-of-the-box by utilizing a structured approach to data storage. This is enabled by the MedRecord class, which organizes data of any complexity within a graph structure. With its Rust backend implementation, MedRecord guarantees high performance, even when working with extremely large datasets.
import medmodels as mm
2. Adding Nodes to a MedRecord#
Let’s begin by introducing some sample medical data:
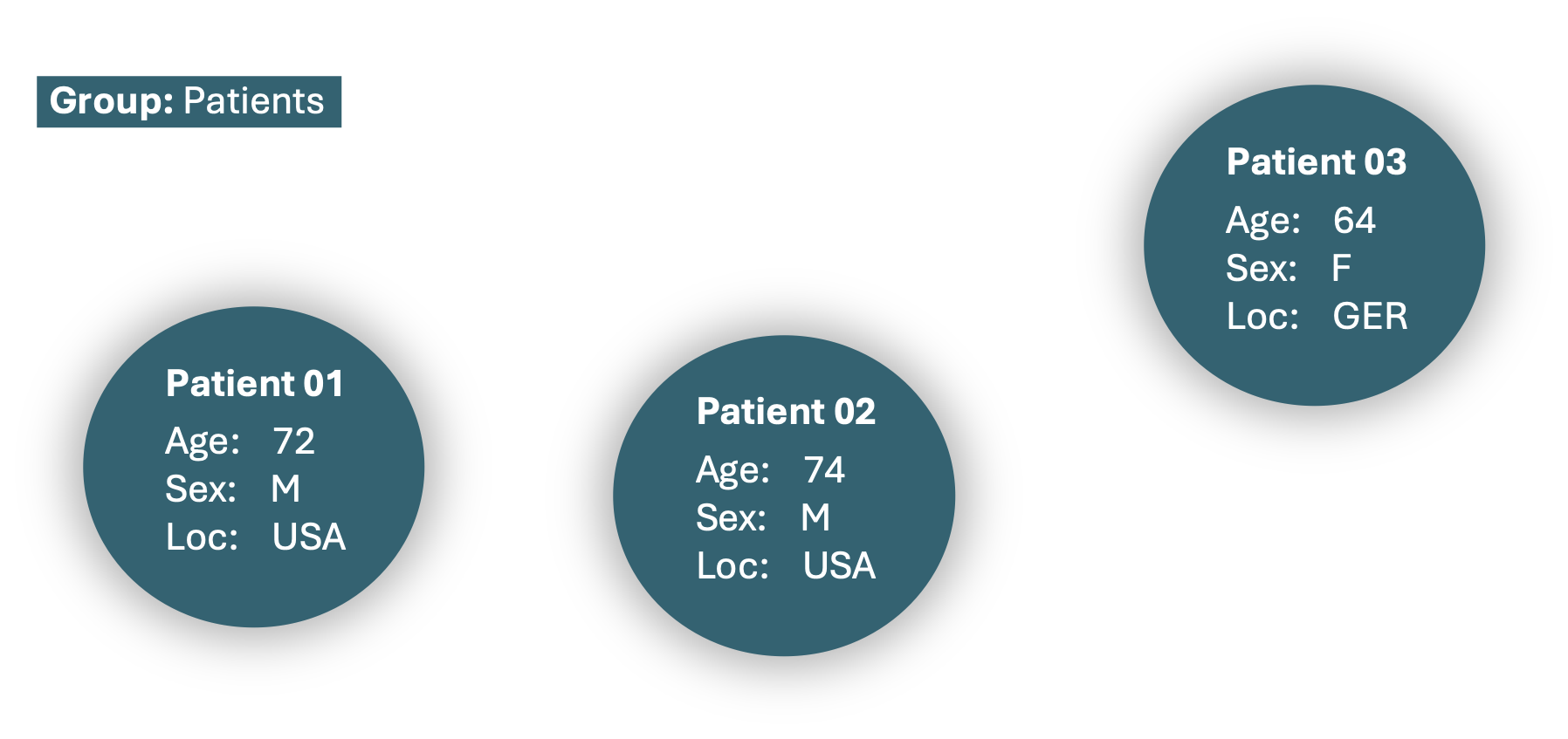
The MedModels MedRecord object, record, now contains three patients. Each patient is identified by a unique index and has specific attributes, such as age, sex, and location. These patients serve as the initial nodes in the graph structure of our MedRecord, and are represented as follows:
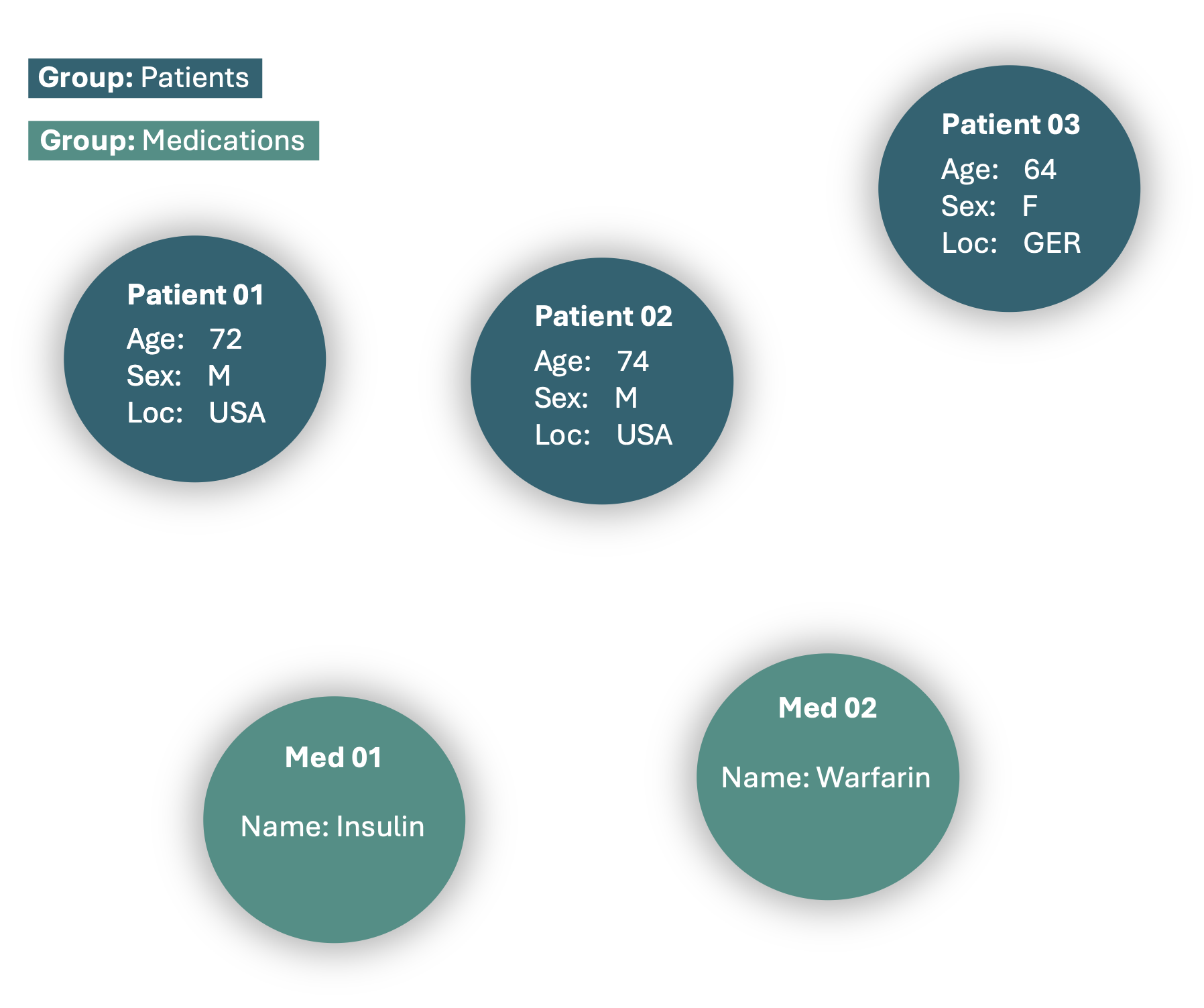
We can now proceed by adding additional data, such as the following medications.
medications = pd.DataFrame(
[["Med 01", "Insulin"], ["Med 02", "Warfarin"]], columns=["ID", "Name"]
)
Using the builder pattern to construct the MedRecord allows us to pass as many nodes and edges as needed. If nodes are not added during the initial graph construction, they can easily be added later to an existing MedRecord by calling add_nodes(), where you provide the DataFrame and specify the column containing the node indices.
record.add_nodes((medications, "ID"), group="Medications")
Methods used in the snippet
add_nodes(): Adds nodes to the MedRecord from different data formats and optionally assigns them to a group.
This will expand the MedRecord, adding several new nodes to the graph. However, these nodes are not yet connected, so let’s establish relationships between them!
Note
Nodes can be added to the MedRecord in a lot of different formats, such as a Pandas DataFrame (as previously shown), but also from a Polars DataFrame:
patient_tuples = [
("Patient 04", {"Age": 45, "Sex": "F", "Loc": "CHI"}),
("Patient 05", {"Age": 26, "Sex": "M", "Loc": "SPA"}),
]
record.add_nodes(patient_tuples, group="Patients")
Or from a NodeTuple:
patient_polars = pl.DataFrame(
[
["Patient 06", 55, "F", "GER"],
["Patient 07", 61, "F", "USA"],
["Patient 08", 73, "M", "CHI"],
],
schema=["ID", "Age", "Sex", "Loc"],
orient="row",
)
record.add_nodes((patient_polars, "ID"), group="Patients")
3. Adding Edges to a MedRecord#
To capture meaningful relationships between nodes, such as linking patients to prescribed medications, we add edges to the MedRecord. These edges must be specified in a relation table, such as the one shown below:
This results in an enlarged Graph with more information.
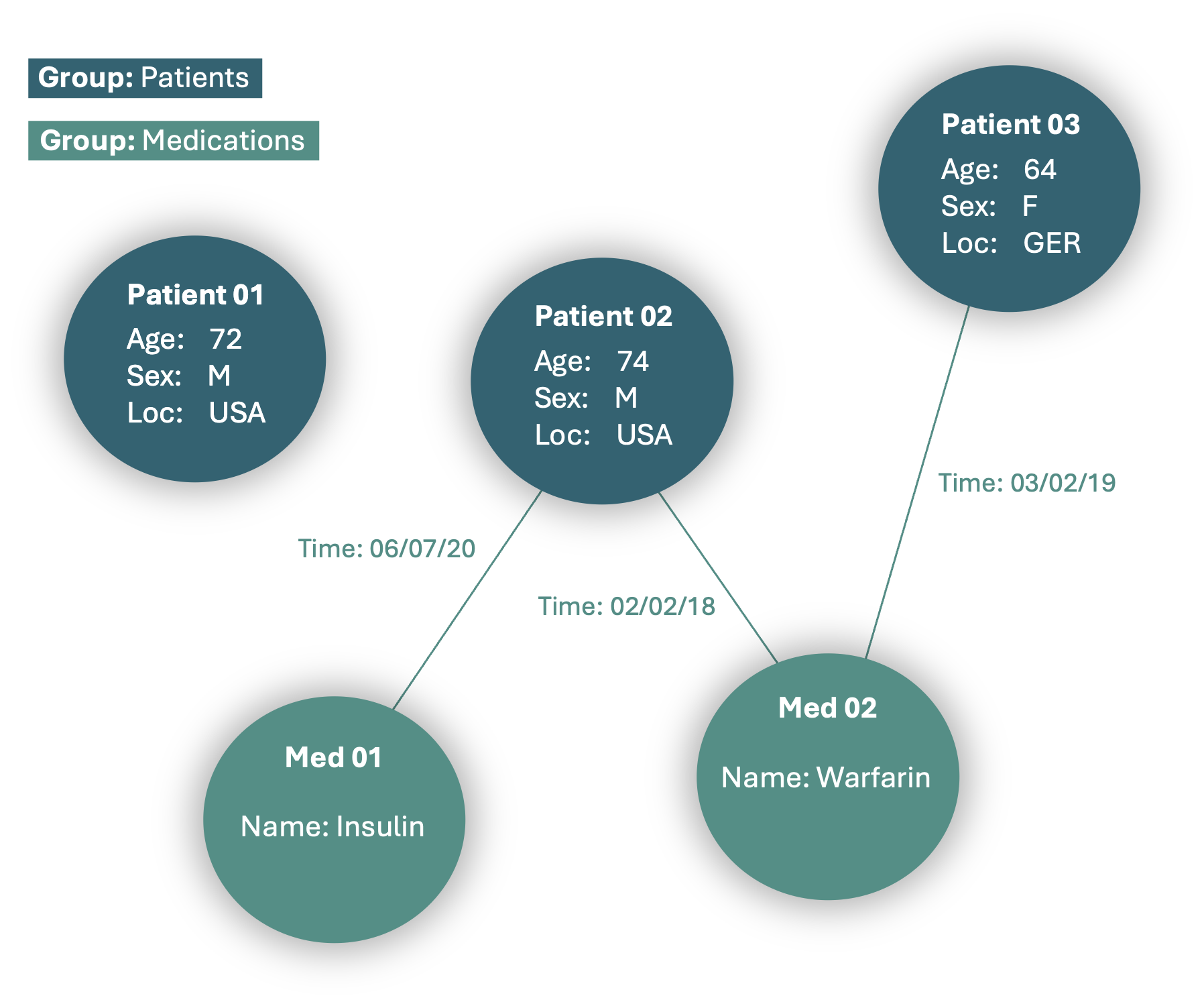
4. Adding Groups to a MedRecord#
For certain analyses, we may want to define specific subcohorts within our MedRecord for easier access. We can do this by defining named groups withing our MedRecord.
record.add_group("US-Patients", nodes=["Patient 01", "Patient 02"])
Methods used in the snippet
add_group(): Adds a group to the MedRecord instance with an optional list of node and/or edge indices.
This group will include all the defined nodes, allowing for easier access during complex analyses. Both nodes and edges can be added to a group, with no limitations on group size. Additionally, nodes and edges can belong to multiple groups without restriction.
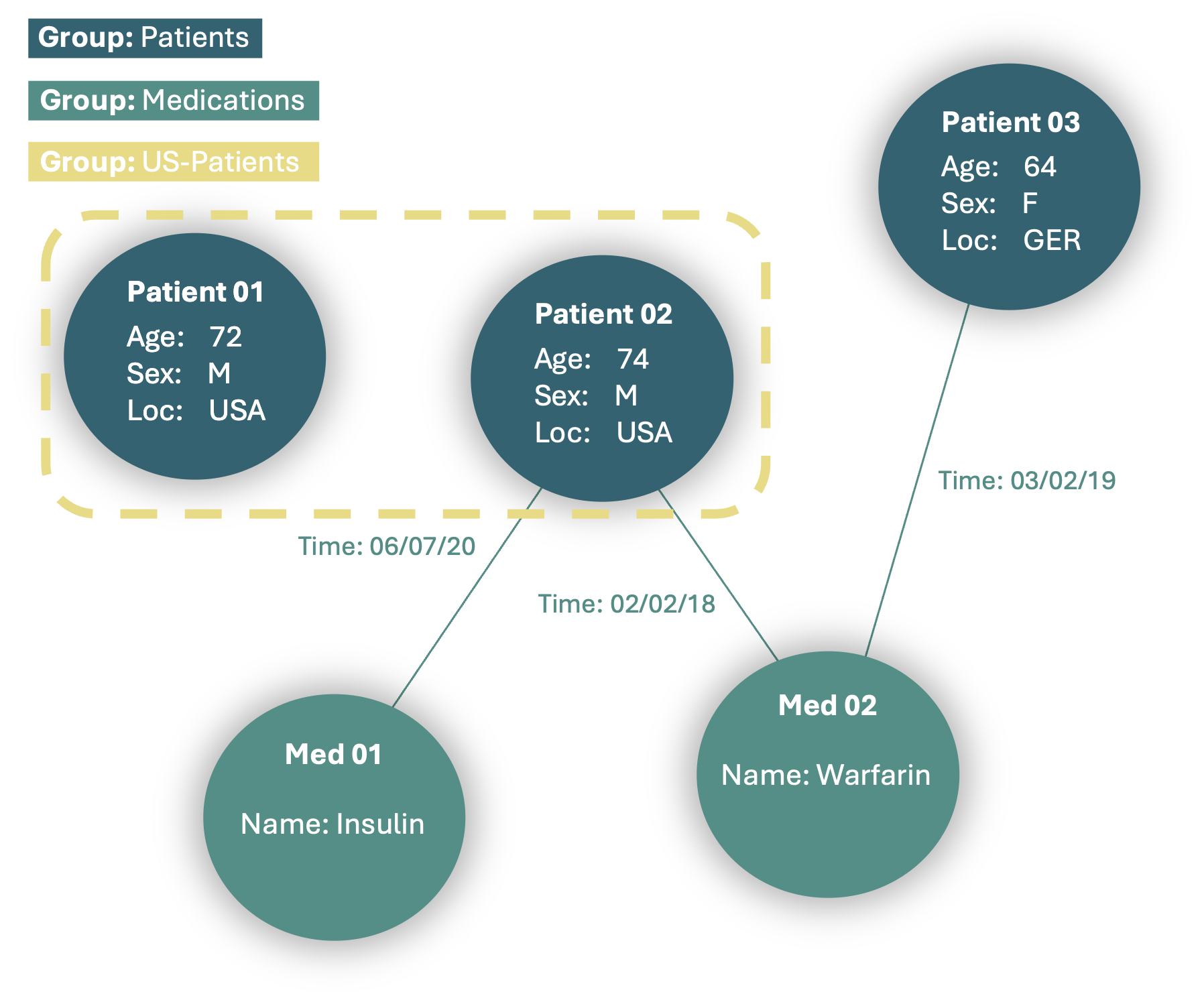
5. Saving and Loading MedRecords#
When building a MedRecord, you may want to save it to create a persistent version. This can be done by storing it as a RON (Rusty Object Notation) file. The MedRecord can then be reloaded, allowing you to create a new instance from the saved RON file.
Methods used in the snippet
to_ron(): Writes the MedRecord instance to a RON file.from_ron(): Creates a MedRecord instance from a RON file.
6. Overview Tables#
The MedRecord class is designed to efficiently handle large datasets while maintaining a standardized data structure that supports complex analysis methods. As a result, the structure within the MedRecord can become intricate and difficult to manage. To address this, MedModels offers tools to help keep track of the graph-based data. One such tool is the overview() method, which prints an overview over all nodes and edges in the MedRecord.
record.overview()
┌──────────────────────────────────────────────────────────────────────────────────────────────────┐
│ Node Overview │
├─────────────┬────────────┬───────────┬────────────────┬────────────────┬─────────────────────────┤
│ Group │ Node Count │ Attribute │ Attribute Type │ Data Type │ Details │
├─────────────┼────────────┼───────────┼────────────────┼────────────────┼─────────────────────────┤
│ │ │ Loc │ Unstructured │ Option[String] │ Distinct value count: 1 │
│ │ ├───────────┼────────────────┼────────────────┼─────────────────────────┤
│ │ │ Name │ Unstructured │ Option[String] │ Distinct value count: 0 │
│ │ ├───────────┼────────────────┼────────────────┼─────────────────────────┤
│ Ungrouped │ 1 │ Sex │ Unstructured │ Option[String] │ Distinct value count: 1 │
│ │ ├───────────┼────────────────┼────────────────┼─────────────────────────┤
│ │ │ │ │ │ Min: 65 │
│ │ │ Age │ Continuous │ Option[Int] │ Mean: 65 │
│ │ │ │ │ │ Max: 65 │
├─────────────┼────────────┼───────────┼────────────────┼────────────────┼─────────────────────────┤
│ Medications │ │ Name │ Unstructured │ String │ Distinct value count: 2 │
├─────────────┤ ├───────────┼────────────────┼────────────────┼─────────────────────────┤
│ │ │ │ │ │ Min: 72 │
│ │ │ Age │ Continuous │ Int │ Mean: 73 │
│ │ 2 │ │ │ │ Max: 74 │
│ US-Patients │ ├───────────┼────────────────┼────────────────┼─────────────────────────┤
│ │ │ Loc │ Unstructured │ String │ Distinct value count: 1 │
│ │ ├───────────┼────────────────┼────────────────┼─────────────────────────┤
│ │ │ Sex │ Unstructured │ String │ Distinct value count: 1 │
├─────────────┼────────────┼───────────┼────────────────┼────────────────┼─────────────────────────┤
│ │ │ Loc │ Unstructured │ String │ Distinct value count: 4 │
│ │ ├───────────┼────────────────┼────────────────┼─────────────────────────┤
│ │ │ │ │ │ Min: 26 │
│ Patients │ 8 │ Age │ Continuous │ Int │ Mean: 58.75 │
│ │ │ │ │ │ Max: 74 │
│ │ ├───────────┼────────────────┼────────────────┼─────────────────────────┤
│ │ │ Sex │ Unstructured │ String │ Distinct value count: 2 │
└─────────────┴────────────┴───────────┴────────────────┴────────────────┴─────────────────────────┘
┌───────────────────────────────────────────────────────────────────────────────────────────────────┐
│ Edge Overview │
├───────────┬────────────┬───────────┬────────────────┬──────────────────┬──────────────────────────┤
│ Group │ Edge Count │ Attribute │ Attribute Type │ Data Type │ Details │
├───────────┼────────────┼───────────┼────────────────┼──────────────────┼──────────────────────────┤
│ Ungrouped │ 3 │ Date │ Temporal │ Option[DateTime] │ Min: 2018-02-02 00:00:00 │
│ │ │ │ │ │ Max: 2020-06-07 00:00:00 │
└───────────┴────────────┴───────────┴────────────────┴──────────────────┴──────────────────────────┘
Methods used in the snippet
overview(): Gets a summary for all nodes and edges in groups and their attributes.
7. Accessing Elements in a MedRecord#
Now that we have stored some structured data in our MedRecord, we might want to access certain elements of it. The main way to do this is by either selecting the data with their indices or via groups that they are in.
We can, for example, get all available nodes:
record.nodes
['Med 02', 'Patient 03', 'Patient 04', 'Patient 05', 'Patient 01', 'Patient 07', 'Patient 02', 'Med 01', 'Patient 08', 'Patient 09', 'Patient 06']
Or access the attributes of a specific node:
record.node["Patient 01"]
{'Age': 72, 'Sex': 'M', 'Loc': 'USA'}
Or a specific edge:
record.edge[0]
{'Date': datetime.datetime(2020, 6, 7, 0, 0)}
Or get all available groups:
record.groups
['Medications', 'Patient-Medication', 'Patients', 'US-Patients']
Or get all that nodes belong to a certain group:
record.nodes_in_group("Medications")
['Med 02', 'Med 01']
Methods used in the snippet
nodes: Lists the node indices in the MedRecord instance.node[]: Provides access to node information within the MedRecord instance via an indexer, returning a dictionary with node indices as keys and node attributes as values.edge[]: Provides access to edge attributes within the MedRecord via an indexer, returning a dictionary with edge indices and edge attributes as values.groups(): Lists the groups in the MedRecord instance.nodes_in_group(): Retrieves the node indices associated with the specified group(s) in the MedRecord.
The MedRecord can be queried in very advanced ways in order to find very specific nodes based on time, relations, neighbors or other. These advanced querying methods are covered in one of the next sections of the user guide, Query Engine.
8. Full example Code#
The full code examples for this chapter can be found here:
import pandas as pd
import polars as pl
import medmodels as mm
# Patients DataFrame (Nodes)
patients = pd.DataFrame(
[
["Patient 01", 72, "M", "USA"],
["Patient 02", 74, "M", "USA"],
["Patient 03", 64, "F", "GER"],
],
columns=["ID", "Age", "Sex", "Loc"],
)
# Medications DataFrame (Nodes)
medications = pd.DataFrame(
[["Med 01", "Insulin"], ["Med 02", "Warfarin"]], columns=["ID", "Name"]
)
# Patients-Medication Relation (Edges)
patient_medication = pd.DataFrame(
[
["Patient 02", "Med 01", pd.Timestamp("20200607")],
["Patient 02", "Med 02", pd.Timestamp("20180202")],
["Patient 03", "Med 02", pd.Timestamp("20190302")],
],
columns=["Pat_ID", "Med_ID", "Date"],
)
record = mm.MedRecord.builder().add_nodes((patients, "ID"), group="Patients").build()
record.add_nodes((medications, "ID"), group="Medications")
patient_tuples = [
("Patient 04", {"Age": 45, "Sex": "F", "Loc": "CHI"}),
("Patient 05", {"Age": 26, "Sex": "M", "Loc": "SPA"}),
]
record.add_nodes(patient_tuples, group="Patients")
patient_polars = pl.DataFrame(
[
["Patient 06", 55, "F", "GER"],
["Patient 07", 61, "F", "USA"],
["Patient 08", 73, "M", "CHI"],
],
schema=["ID", "Age", "Sex", "Loc"],
orient="row",
)
record.add_nodes((patient_polars, "ID"), group="Patients")
record.add_edges((patient_medication, "Pat_ID", "Med_ID"))
record.add_group("US-Patients", nodes=["Patient 01", "Patient 02"])
record.add_nodes(
(
pd.DataFrame(
[["Patient 09", 65, "M", "USA"]], columns=["ID", "Age", "Sex", "Loc"]
),
"ID",
),
)
record.overview()
# Adding edges to a certain group
record.add_group("Patient-Medication", edges=record.edges)
# Getting all available nodes
record.nodes
# Accessing a certain node
record.node["Patient 01"]
# Accessing a certain edge
record.edge[0]
# Getting all available groups
record.groups
# Getting the nodes that are within a certain group
record.nodes_in_group("Medications")
record.to_ron("record.ron")
new_record = mm.MedRecord.from_ron("record.ron")